Performance¶
This page reports performance measurements on Asap on the Niflheim cluster here at DTU. To make it easy to estimate the running time of your own scripts, all times are given in microseconds per atom per timestep. So if you want to do N timesteps with M atoms, your running time will be X*N*M*1e-6 seconds.
Computers¶
Timing has been done on the following computers:
- xeon16
16-core node with Intel Ivy Bridge cpus (Intel(R) Xeon(R) CPU E5-2650 v2 @ 2.60GHz)
- xeon24
24-core node with Intel Broadwell cpus (Intel(R) Xeon(R) CPU E5-2650 v4 @ 2.20GHz).
- xeon40_clx
40-core node with Intel Cascade Lake cpus (Intel(R) Xeon(R) Gold 6242R CPU @ 3.10GHz)
- xeon56
56-core node with Intel Icelake cpus (Intel(R) Xeon(R) Gold 6348 CPU @ 2.60GHz)
Except where otherwise noted, Asap was compiled using the EasyBuild
foss/2023b toolchain, which contains the GNU GCC compiler suite
version 10.2.0.
Serial simulations¶
The tests were performed as single cores jobs. All cores were running the same script (to give realistic performance for a busy high-performance cluster), and the times are averages over all the separate jobs at each core.
Four kinds of simulations were run, with varying number of atoms. The system was a Silver-Gold random alloy in the FCC structure, unless otherwise noted. Interactions were described with the standard EMT potential provided by Asap.
- Verlet
Velocity Verlet dynamics at 300 K. The simplest possible type of simulation.
- Verlet (single element)
As above, but with a pure silver system. A number of optimizations are enabled automatically in the EMT potential if the system consists of only a single chemical element.
- Langevin (300K)
Langevin dynamics (NVT ensemble) at 300K.
- Langevin (2000K)
Langevin dynamics at 2000K. This gives more frequent neighbor list updates, and in parallel simulations (see below) it will also result in frequent migrations between cores.
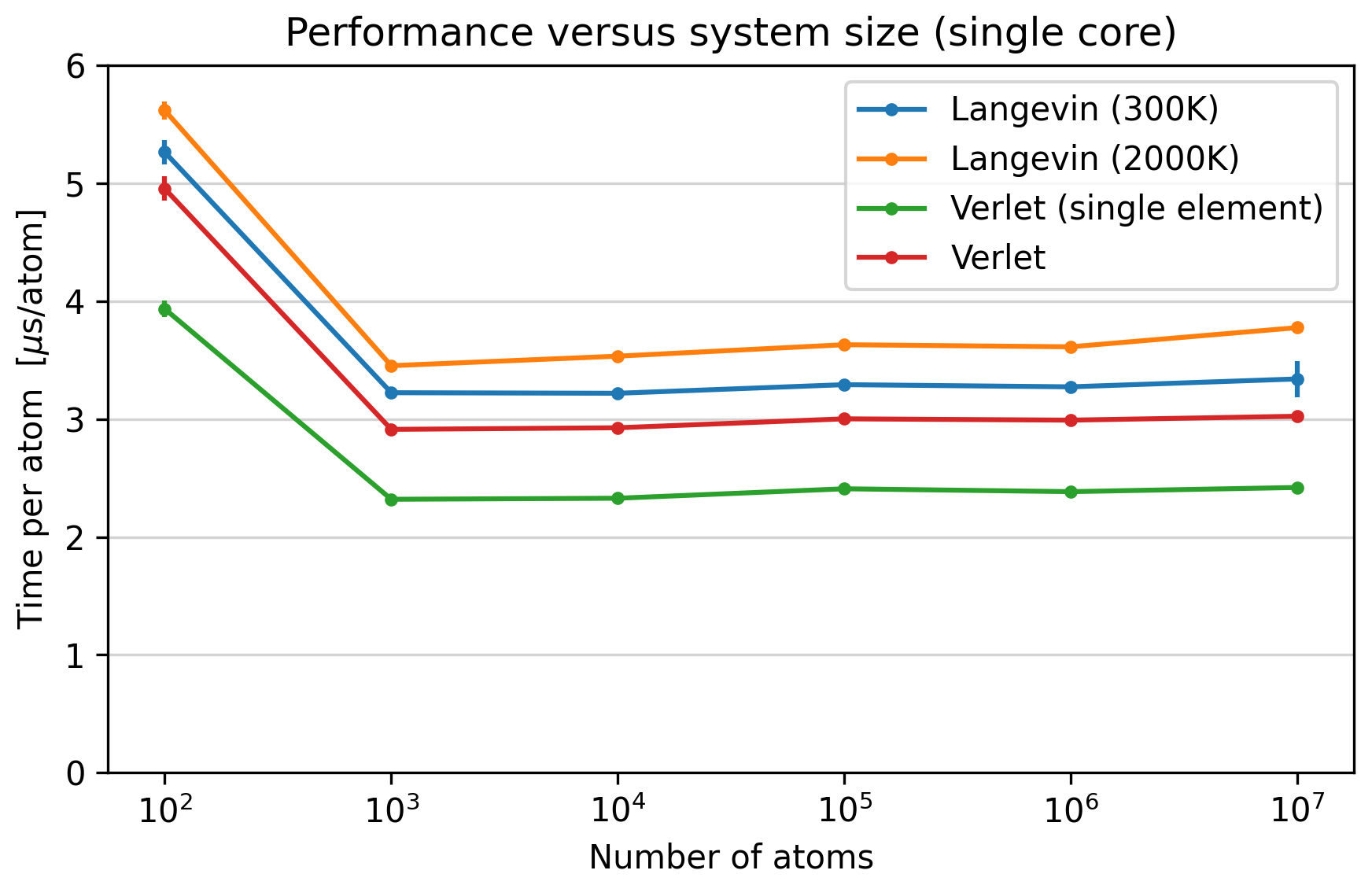
Performance versus system size for the four different
types of simulations. All data are taken on the
xeon40_clx nodes at Niflheim. Asap was compiles with the
EasyBuild foss/2023b toolchain.¶
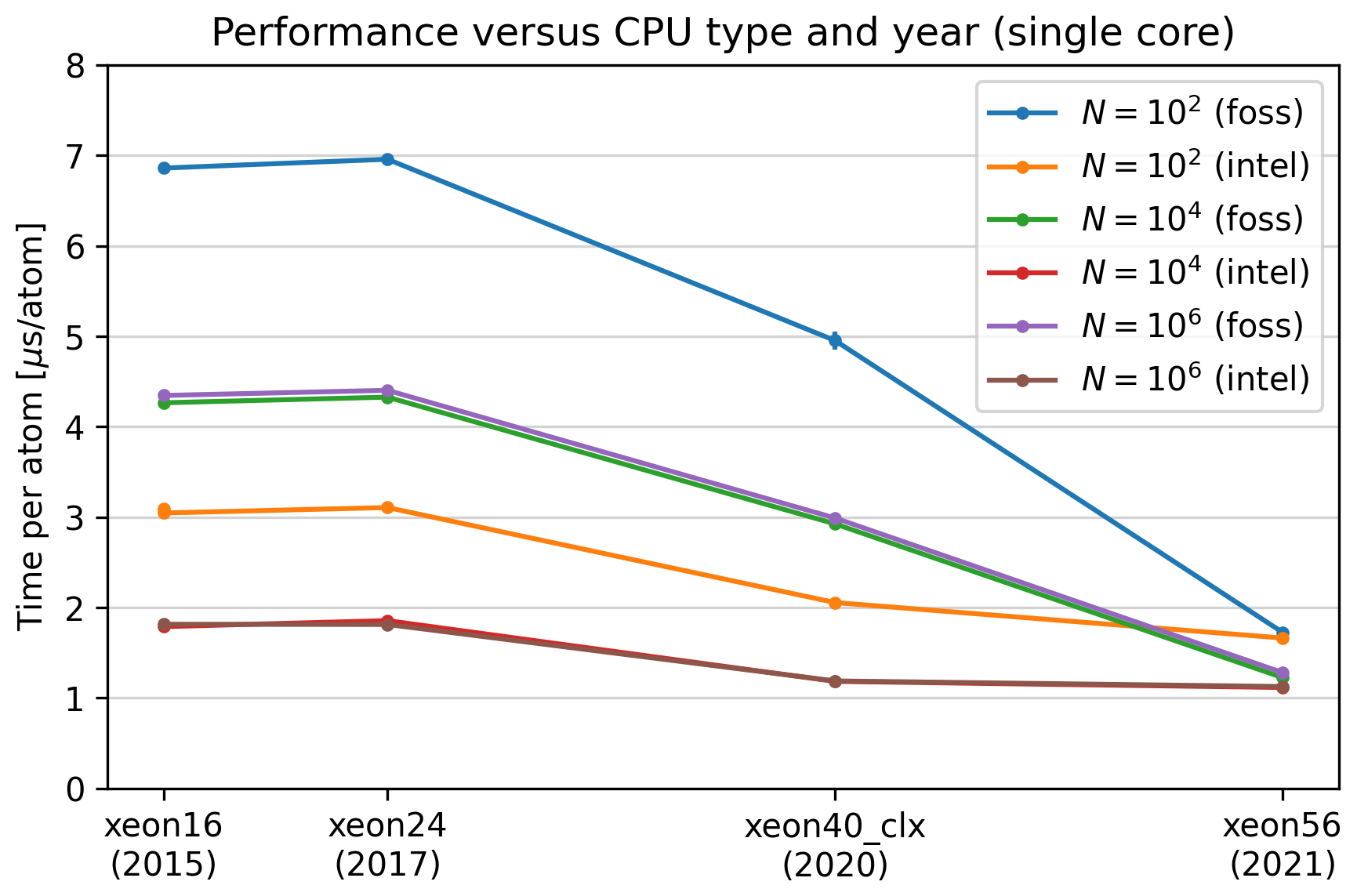
Performance for the different kinds of compute nodes in
the DTU Niflheim system. The year in parentesis is the
installation year. Asap was compiled with two different
toolchains, the EasyBuild foss/2023b (marked ‘foss’)
and intel/2023b (marked ‘intel’). The simulation type
was ‘Verlet’ as described above. The red curve is
almost invisible as it lies behind the brown curve.¶
Note that for all CPUS except the newest, the Intel compiler suite gives a factor two in performance. For the newest CPUs, there is no difference. It looks like it is the foss compilers that are catching up in this case.
Multi-threaded simulations¶
The tests were performed on a single xeon40_clx node with 40 cores. When multithreading on N cores, other cores were kept busy by simultaneously running 40/N such jobs (in some cases leaving a few cores empty).
Below is reported the time per atom for a single timestep, and also the parallel efficiency. A parallel efficiency of 1.0 would correspond to a time step being a factor N faster on N cores.
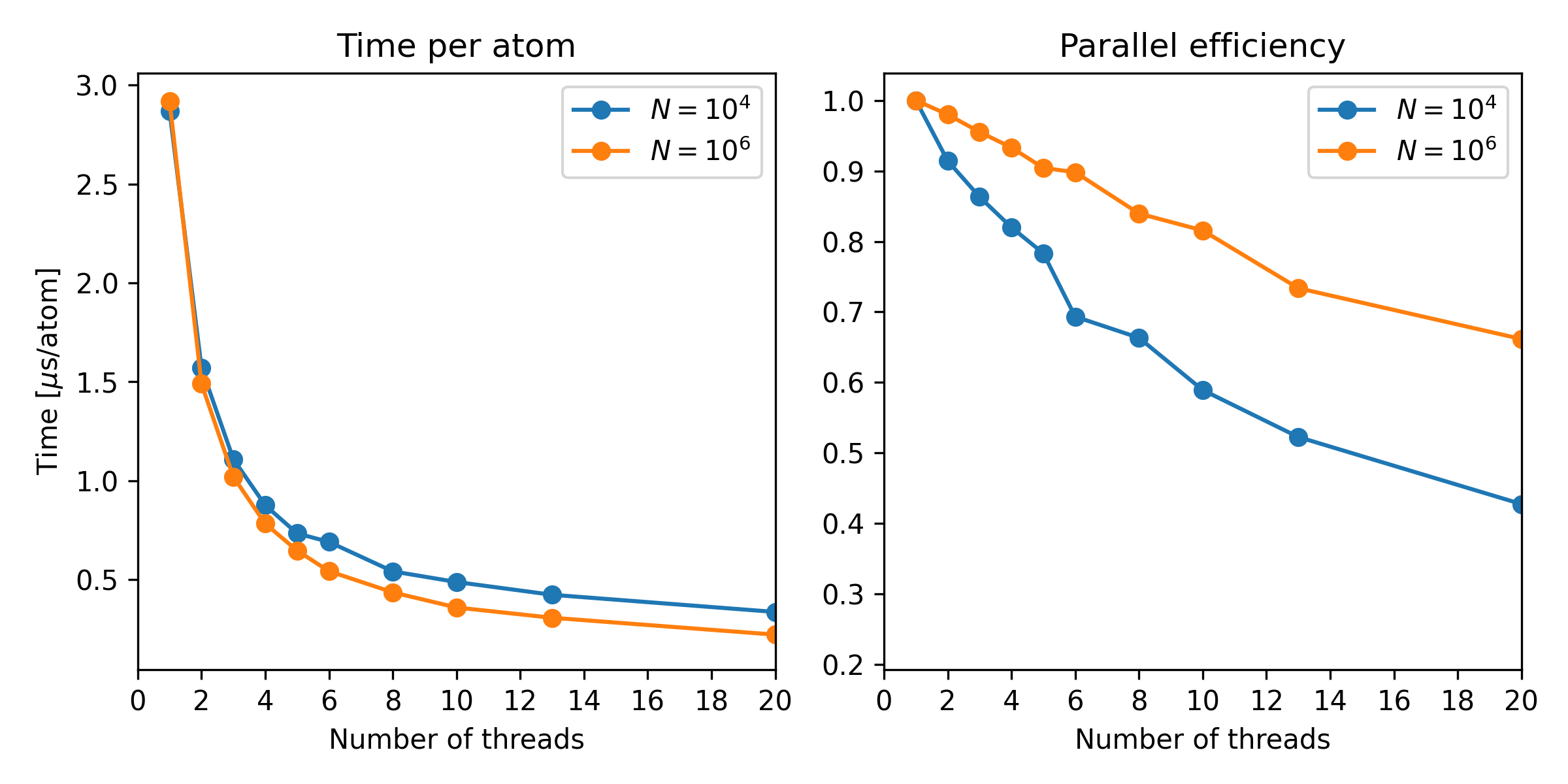
Performance versus number of threads for the default Verlet simulation. All data are taken on the xeon40_clx nodes at Niflheim. Simulations were done with two different system sizes, 10000 and a million atoms.¶
Parallel performance (MPI parallelization)¶
The tests were performed on up to 10 xeon40_clx nodes and 10 xeon56 nodes,
with 40 and 56 cpu cores respectively. Asap was compiled with the
foss/2023a toolchain, and the Verlet simulation test. Two data sets
were taken, with 100,000 and 10,000,000 atoms per core. The largest test
thus had 5.6 billion atoms (simulations with more than 2 billion atoms
require Asap version 3.13.2; more than 4 billion require 3.12.3).
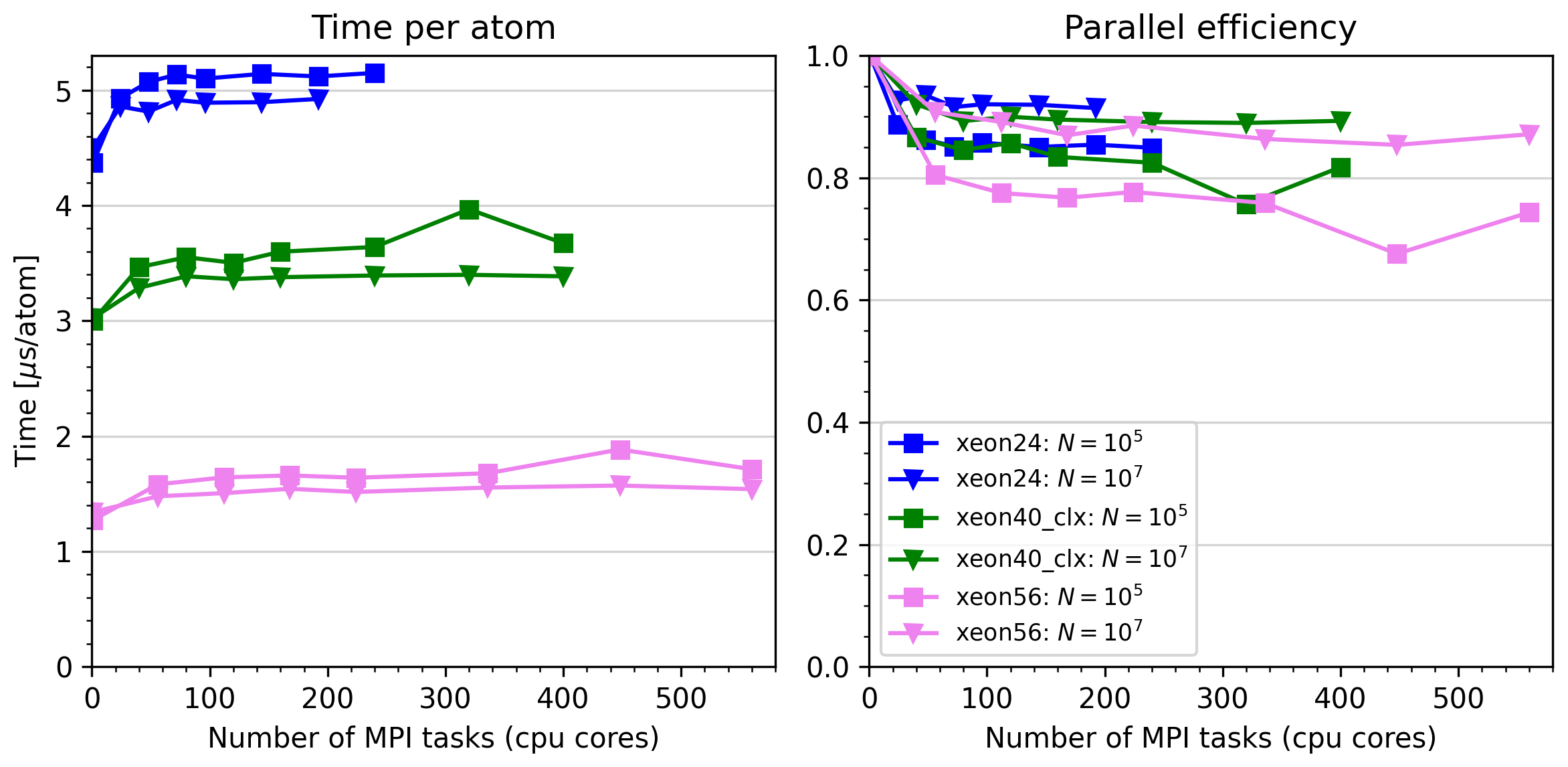
Performance versus number of cpu cores (MPI tasks) for the default Verlet simulations. It is seen that Asap parallelizes with more than 85% efficiency on more than 500 cores, provided the simulations are sufficiently large.¶